Karozis S., Kanourgiakis M.
The in-silico studies reported so far for the representation of the structure and the evaluation of the transport properties of lipid bilayers are in general based on assumptions and approaches that simplify the real system and problem. Nevertheless, the structure and organization of the lipid bilayers strongly affect transport coefficients. This is a quite important observation, showing that simulations can be meaningful only when addressing realistic structures, mimicking the actual lipid phase system as elaborately as possible.
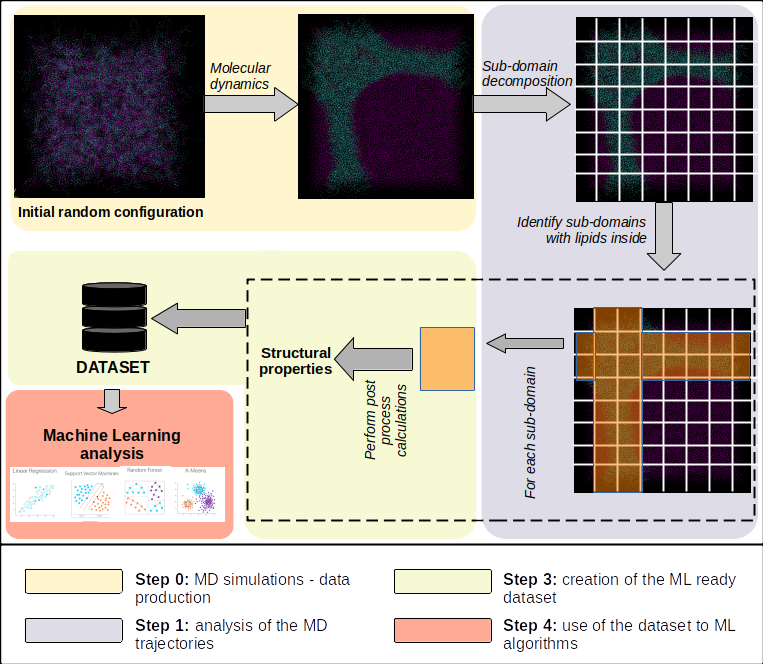
In the current study, a computational tool is presented that uses Molecular Dynamics simulations (MD) results of spontaneous self-assembly lipid bilayer structures with different oriented and shaped lipid bilayer, in order to analyze the resulted trajectories, creating a Machine Learning (ML) ready dataset that can be used in a series of ML algorithms, depending the case. The development of the tool is in the alpha stage, where tests are performed, with a planned public release in free and open source. The tool’s development state is an alpha version, where tests and debugging are performed. As future work, the outcome and results of the a case study is planned to be used, alongside the first public release of the code under free and open source license. The tool is hosted at: https://mssg.ipta.demokritos.gr/gitlab/skarozis/toobba
AI in Natural Sciences and Technology (AINST2020) proceeding
This research is co-financed by Greece and the European Union (European Social Fund - ESF) through the Operational Programme «Human Resources Development, Education and Lifelong Learning» in the context of the project "Reinforcement of Postdoctoral Researchers - 2nd Cycle" (MIS-5033021), implemented by the State Scholarships Foundation (IKY).